Protein NMR Projects
Nuclear Magnetic Resonance Spectroscopy - From Nuclear Spin Interactions To Molecular Structure
Our group is interested in the development of solid-state Nuclear Magnetic Resonance (NMR) methods for the characterization of molecular structure, dynamics and interactions at atomic resolution. Our interdisciplinary research spans a wide range of areas from quantum mechanics of spin interactions to practical applications:
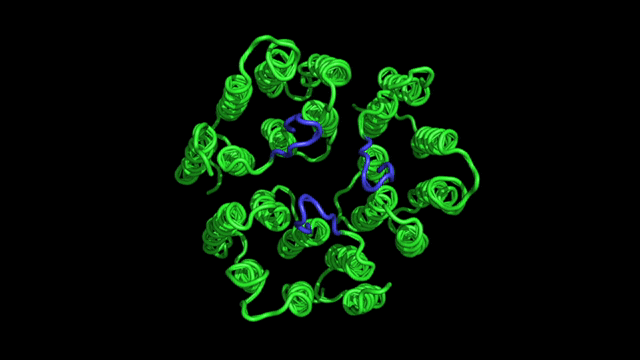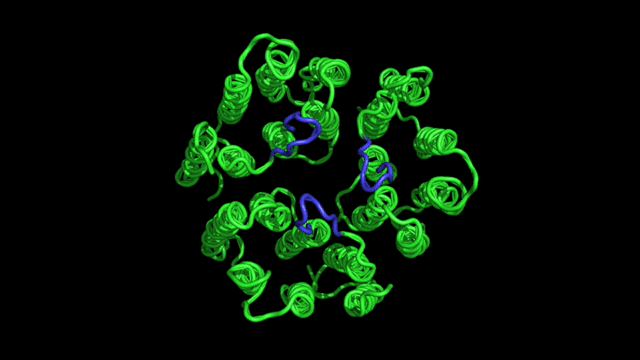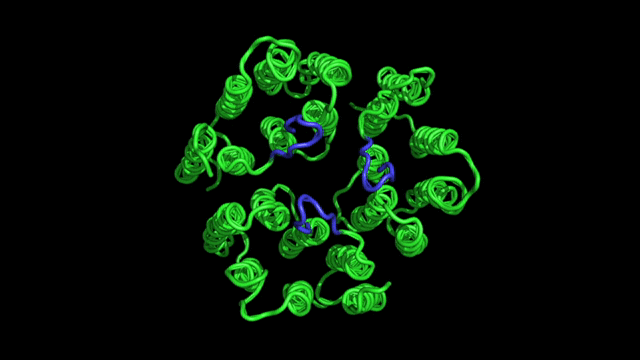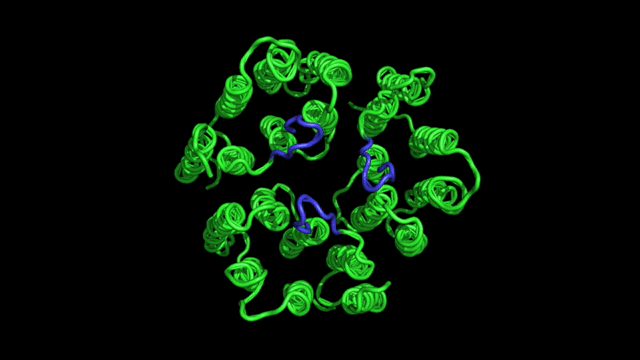
- Designing new experiments for determination of protein structure. A typical NMR experiment consists of a sequence of radio-frequency pulses, which can be designed to rotate spins and to tailor spin interactions to an optimal form. We are interested in developing new methods for specific experimental needs.
- Protein dynamics. In an NMR experiment, nuclear spins are perturbed from equilibrium by radio-frequency pulses; they will eventually come back to equilibrium at a rate that depends on the molecular motions. From the analysis of nuclear spin relaxation rates one can derive both the time scales and amplitudes of motions. This information is directly related to how molecules function.
- Applications of NMR in biophysics and biomedicine. We apply these methods to problems of general biophysical interest, as well as medical/biotechnological relevance, e.g., design of new pharmacological drugs, understanding of the molecular origins of diseases, etc.

Graduate student positions available
We are seeking MSc and PhD students to work in our group. Current projects in the lab focus on understanding of membrane protein folding and protein dynamics; on the role of protein aggregation in Parkinson’s Disease.
We have the equipment and expertise and need your talent and enthusiasm.
If interested, do not hesitate to email Vlad Ladizhansky directly, and inquire about current possibilities: vladizha@uoguelph.ca.
Representative publications
- S. Wang et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nature Methods (2013), 10, 107.
- D. Good et al. Solid-state NMR provides evidence for small-amplitude slow domain motions in a multi-spanning transmembrane α–helical protein, J. Am. Chem. Soc. (2017), 139, 9246.
- P. Xiao et al. Solid-state NMR spectroscopy based atomistic view of a membrane protein unfolding pathway. Nature Commun (2019), 10, 3867.